About me
Hello. I am an ORISE postdoctoral fellow at USDA-ARS working with UAV images for maize plant phenotyping. I work with digital images to extract plant traits using computer vision and machine learning techniques. I also work on the development of robotic systems for agriculture and forestry applications. I completed my dual PhD in Engineering and Forestry from North Carolina State University in 2022.
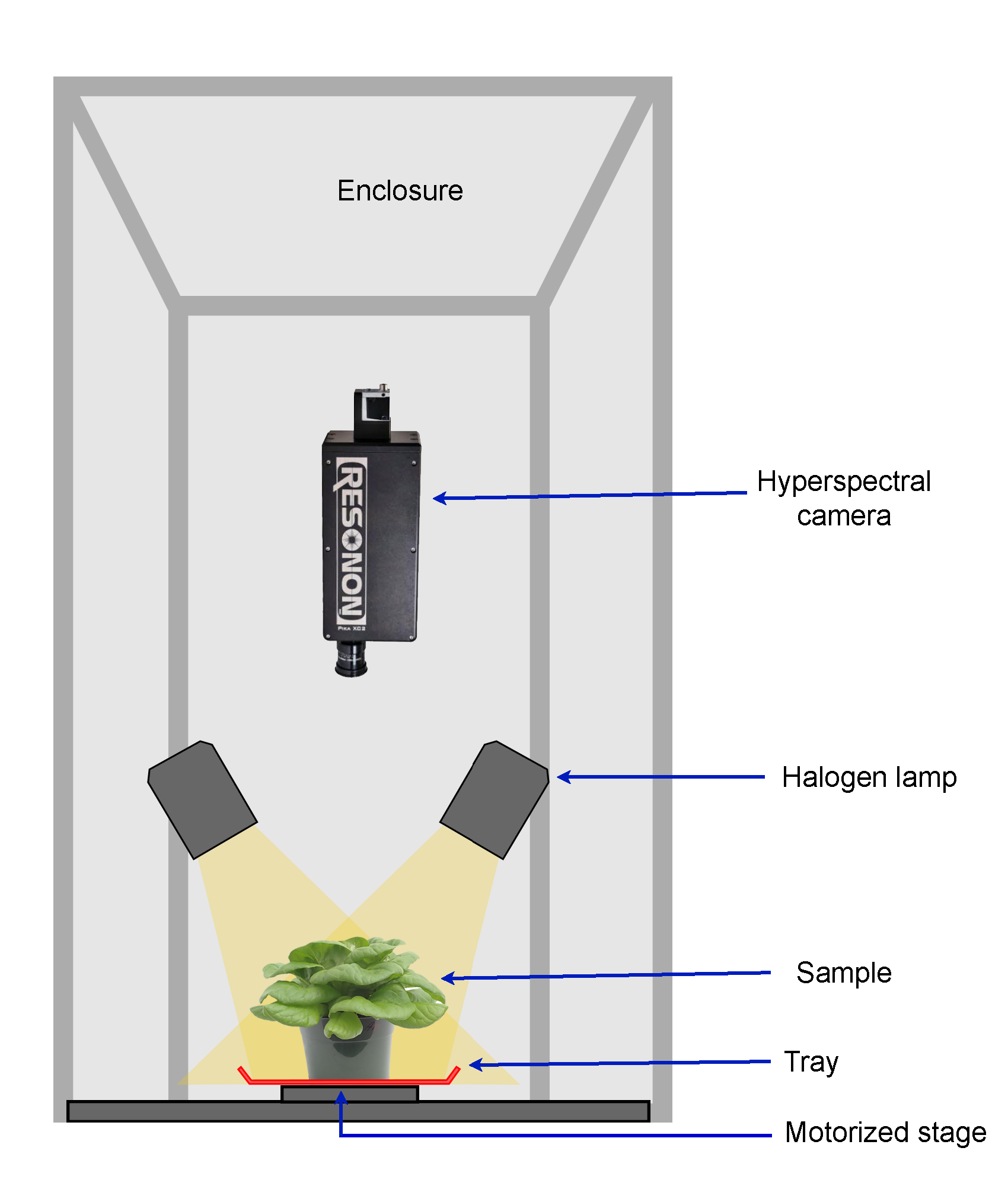
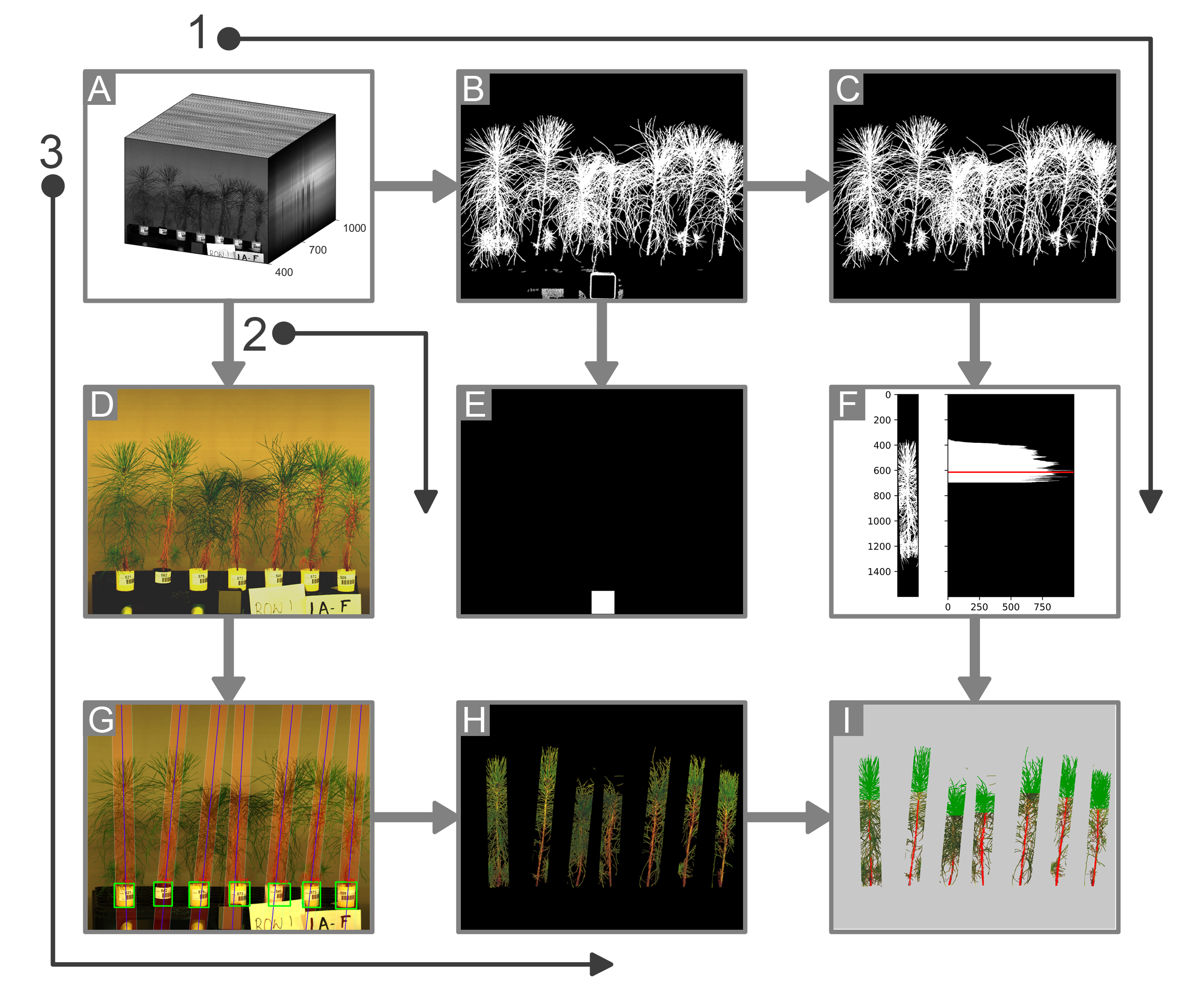
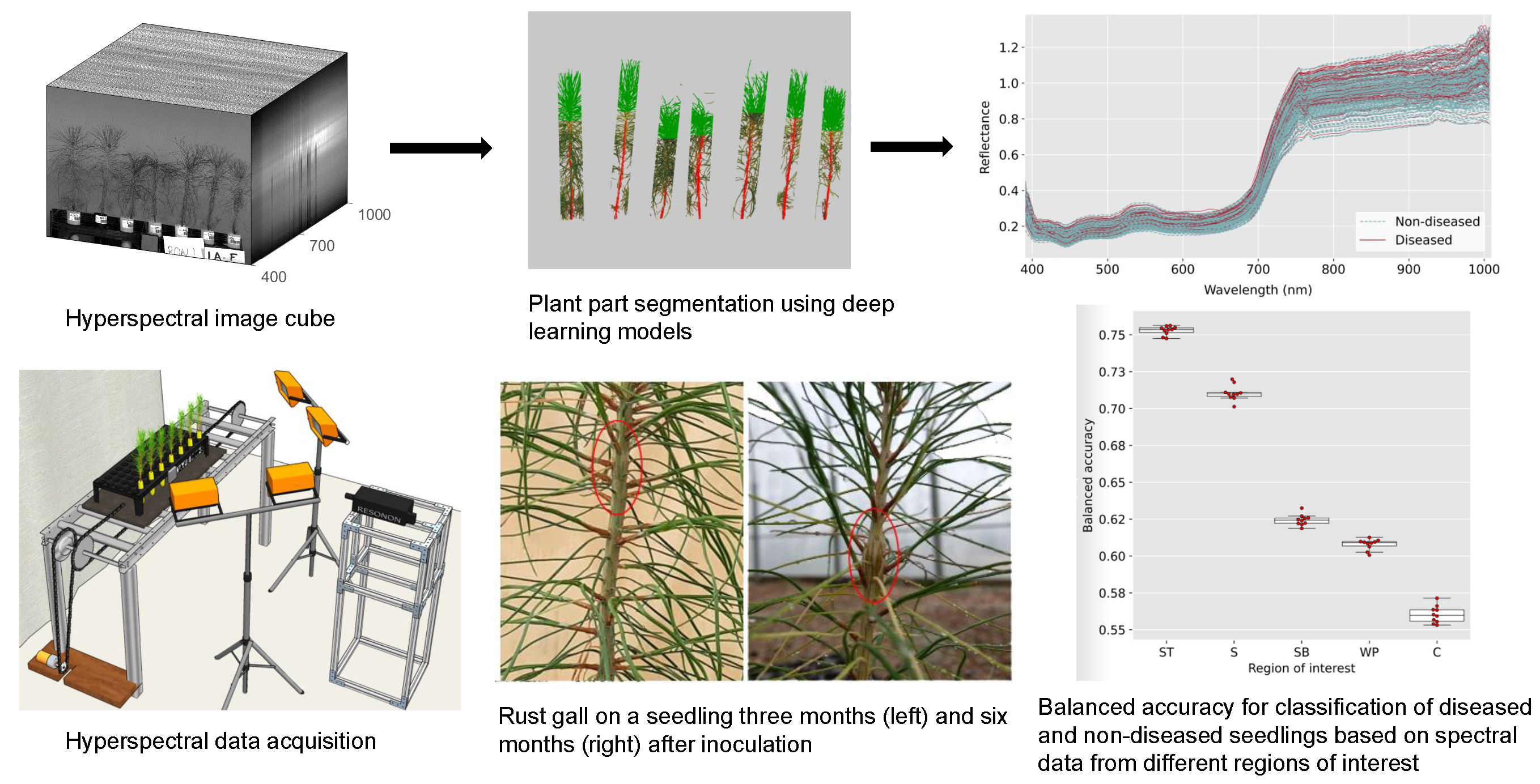
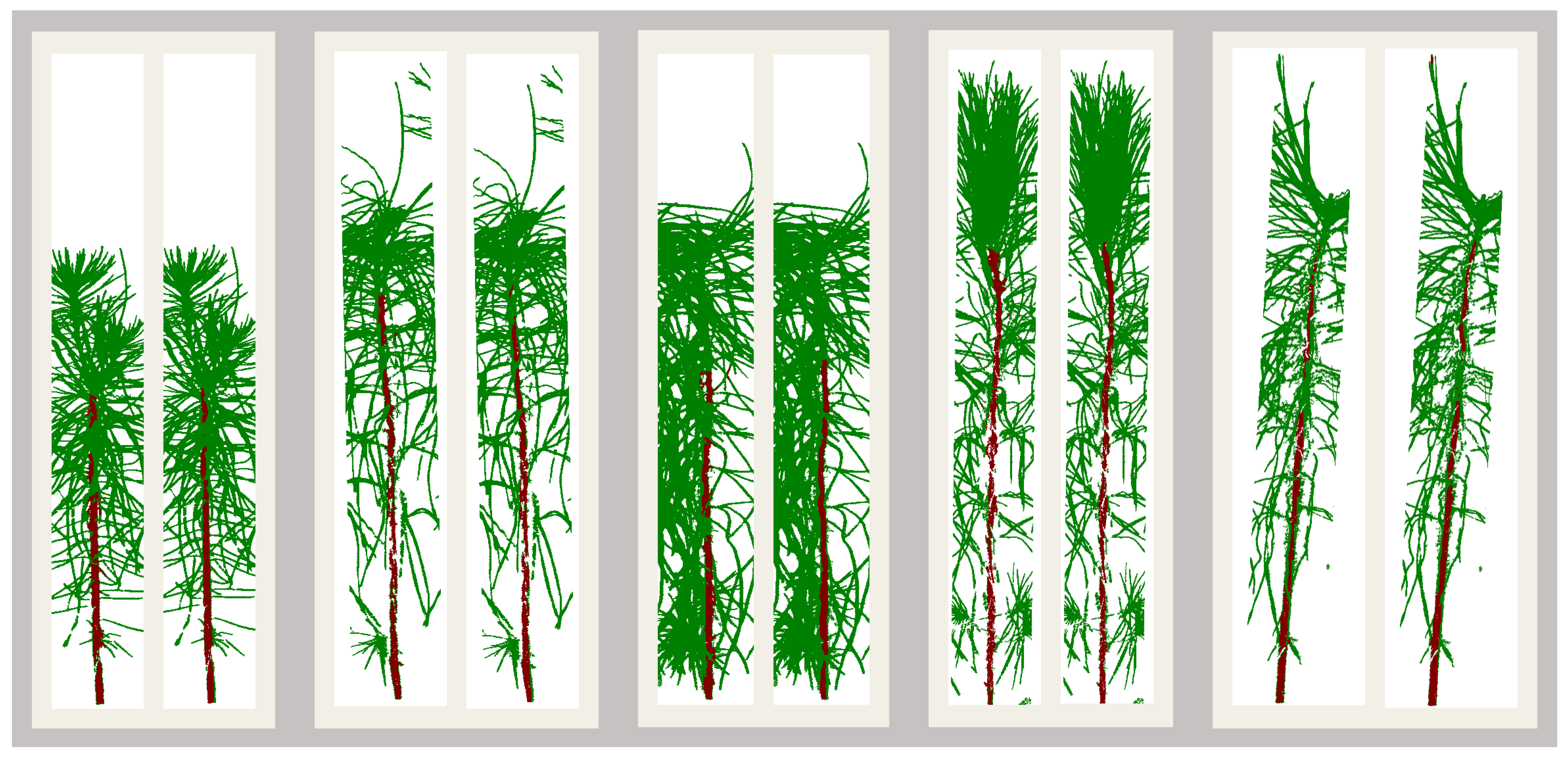
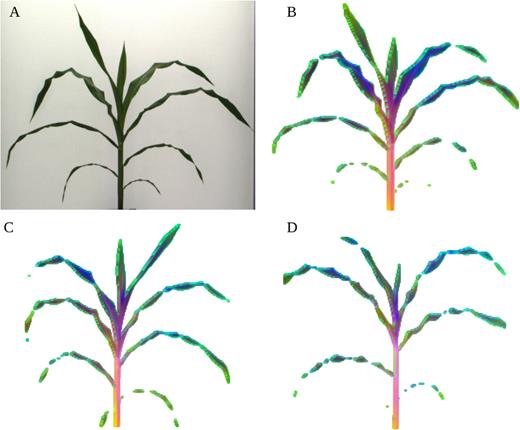
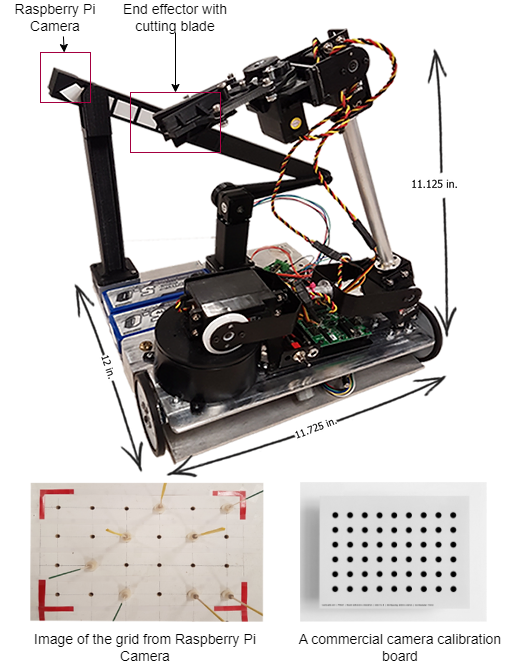
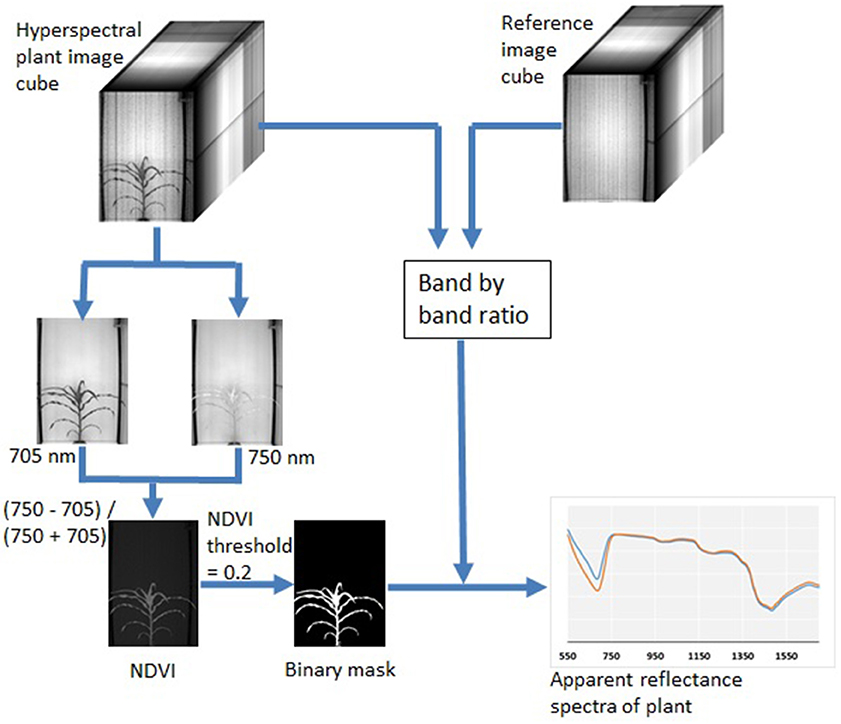
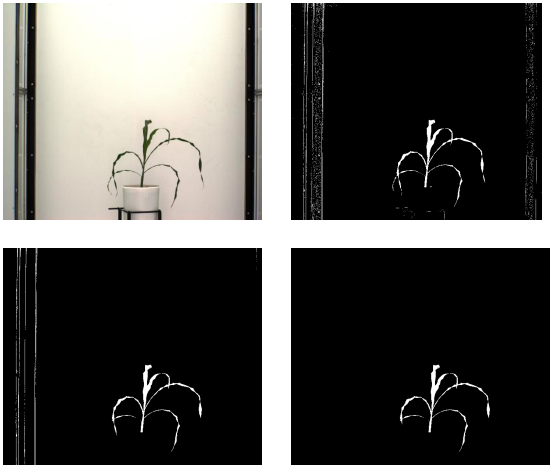
A large part of my work with plant images has been focused on the use of hyperspectral imaging to find optical signals of plant biotic and abiotic stress. At NC State, I developed a robotic pollination system for controlled pollination in loblolly pine seed orchards.
On this page, you will find a brief description of my projects with links for more information. Details on many of these projects are also accessible through my publications page.